Regulation of Phospholipid Metabolism/Signaling in Yeast
Research in our laboratory utilizes molecular genetics and biochemical approaches to study the regulation of phospholipid metabolism in the yeast Saccharomyces cerevisiae. Phospholipids are essential molecules that contribute to the structural definition of cell membranes, and participate in the regulation of cellular processes as signaling molecules and as reservoirs of lipid messengers. Our laboratory has made significant contributions to the understanding of phospholipid synthesis in yeast through the purification and characterization of several enzymes, and through the isolation and characterization of key genes. The laboratory has played a major role in the discovery that the expression of phospholipid synthesis enzymes is regulated by phospholipid precursors (e.g., inositol, choline, ethanolamine, and serine) and the mineral zinc; and that key enzymes are regulated by membrane (e.g., phospholipids, sphingolipids) - and cytosolic (e.g., inositol, serine, ATP, CTP) -associated components, by covalent modifications via protein kinases and protein phosphatases, and by proteasome-mediated degradation. These forms of enzyme regulation have profound effects on membrane phospholipid composition and cell physiology, and have important implications for understanding the molecular basis for lipid-based diseases (e.g., obesity, lipodystrophy, diabetes, and heart disease).
Early in my career, I recognized the importance of using yeast as model eukaryotic organism to study the regulation of phospholipid synthesis. That this single cell organism is easily grown in large quantities has facilitated the purification and characterization of phospholipid synthesis enzymes, and the ease of genetic manipulation has facilitated our work on the molecular characterization of phospholipid synthesis regulation. Importantly, our work with yeast has proven to be relevant to the mechanisms that regulate lipid synthesis in humans (e.g., discovery that human lipin is a phosphatidate (PA) phosphatase enzyme).
Over the past few years, our laboratory has focused on a very important branch point in eukaryotic lipid metabolism where PA is the direct precursor to the anionic phospholipids or is dephosphorylated to diacylglycerol as the precursor to the amine containing phospholipids and to the neutral lipid triacylglycerol. In 2006, we discovered the molecular function of a key fat-regulating protein known as lipin. In a mouse model, lipin deficiency causes a selective loss of body fat (i.e., lipodystrophy) whereas an excess of lipin promotes extra body fat (i.e., obesity). How lipin affects fat metabolism, however, has been an enigma due to lack of information on the molecular function of the protein. Our laboratory discovered that lipin is a PA phosphatase that catalyzes the formation of the fat precursor diacylglycerol, which is the immediate precursor to triacylglycerol. This discovery provided an important clue to understanding the molecular basis for lipodystrophy and obesity in humans. In addition, the PA phosphatase activity of lipin may represent an important pharmaceutical target for the control of body fat in humans.
Since its discovery, we have gone on to show the yeast Pah1 PA phosphatase expression is regulated by nutrients (e.g., inositol, zinc) and growth phase, and have defined the transcription factors involved. On a biochemical level, we discovered that the PA phosphatase enzyme is regulated by phospholipids, sphingoid bases, phosphorylation and dephosphorylation, and by proteasomal degradation. Additional work has shown that PA phosphatase function in vivo is counterbalanced by an unconventional CTP-dependent diacylglycerol kinase activity, an enzyme and its encoding DGK1 gene also discovered in our laboratory.
|
|
|
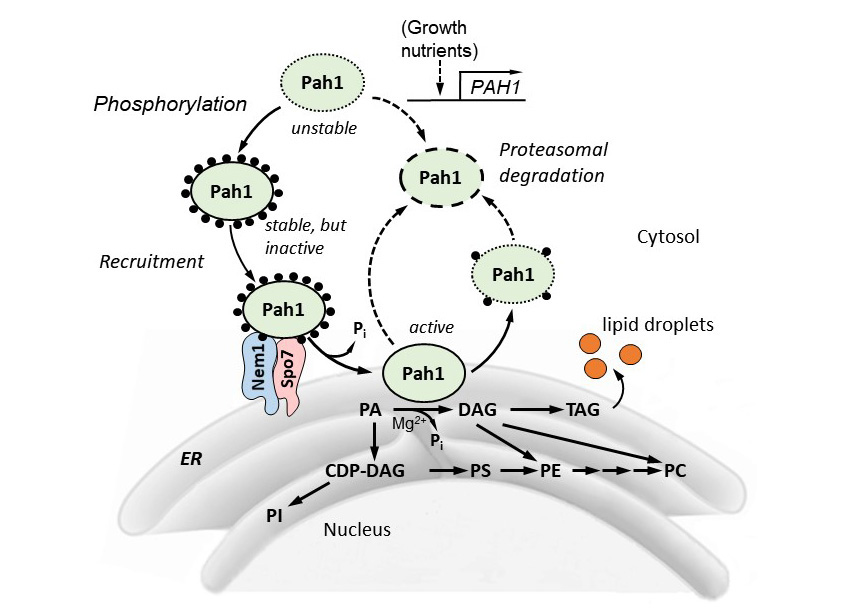
Model for Pah1 recruitment and dephosphorylation by the Nem1-Spo7 protein phosphatase complex. The recruitment and dephosphorylation of the cytosolic-associated phosphorylated (black circles) Pah1 at the nuclear/ER membrane is mediated by its interaction and dephosphorylation by the Nem1-Spo7 protein phosphatase complex. After its association with the membrane, Pah1 binds to and dephosphorylates its substrate PA to form DAG, which is then acylated to TAG that is subsequently stored in cytosolic lipid droplets. PA can also be diverted to the synthesis of phospholipids via CDP-diacylglycerol when Pah1 activity is reduced. Unphosphorylated/dephosphorylated Pah1 is unstable (dotted lines) undergoes proteasomal degradation. Pah1, PAH1-encoded PA phosphatase; PA, phosphatidate; DAG, diaclglycerol; TAG, triacylglycerol; CDP-DAG, cytidine diphosphate diacylglycerol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI, phosphatidylinositol; LDs, lipid droplets.
|